Learning Rmarkdown
- Learn the main components of an Rmarkdown (
.Rmd) document - Understand how to create a new Rmd and add text and images
- Learn how to stitch (or
knit) a document together and share it
Rmarkdown: Knitting Things Together
This entire course website is built using Rmarkdown files. Dissertations, papers, reports, and interactive documents are all very possible using Rmarkdown. While it would be impossible to show you how to do all of these things, the good news is while Rmarkdown is highly customizable, there are only a few key components to learn. We’ll go over some of the main pieces, demonstrate some fun tricks that are made easy using Rmarkdown, and provide a bunch of great resources to help you learn more!
(ref:AHrmdplot) Illustration by @allison_horst

Figure 1: (ref:AHrmdplot)
Reproducible
Depending on the tasks you are faced with, analysis is rarely
something that happens a single time. We are constantly faced with tasks
(sometimes small, sometimes large) that have repetitive information, or
repetitive steps to complete. Making this process more reproducible by
using reusable tools (e.g. {dplyr}) and skills
(R/Rmarkdown!) means you save your future self time and
energy, as well as make it easier to communicate and share your
work.
(ref:AHreproducible) Illustrations from the Openscapes blog Tidy Data for reproducibility, efficiency, and collaboration by Julia Lowndes and Allison Horst”

Figure 2: (ref:AHreproducible)
Combine Text + Code + Figures
Rmarkdown documents may initially seem scary and a bit overwhelming, but once we understand the 3 critical components of an Rmd document, things get easier. There are many flavors and options to customize each of these, but let’s cover the basics.
Rmarkdown Components
There are three main parts of an Rmarkdown
document:(yaml, text, and
code). The first of the three is required no matter
what. The other two, text and
code are more flexible and are not
necessarily required.
RStudio provides an excellent Rmarkdown editor, and even better, the
most recent version of this software provides a nice Visual
Editor mode which makes it even easier to learn the basics of
Markdown and Rmarkdown. Let’s create a new Rmarkdown
document in RStudio.
- File > New File > R Markdown
We should see something like this:
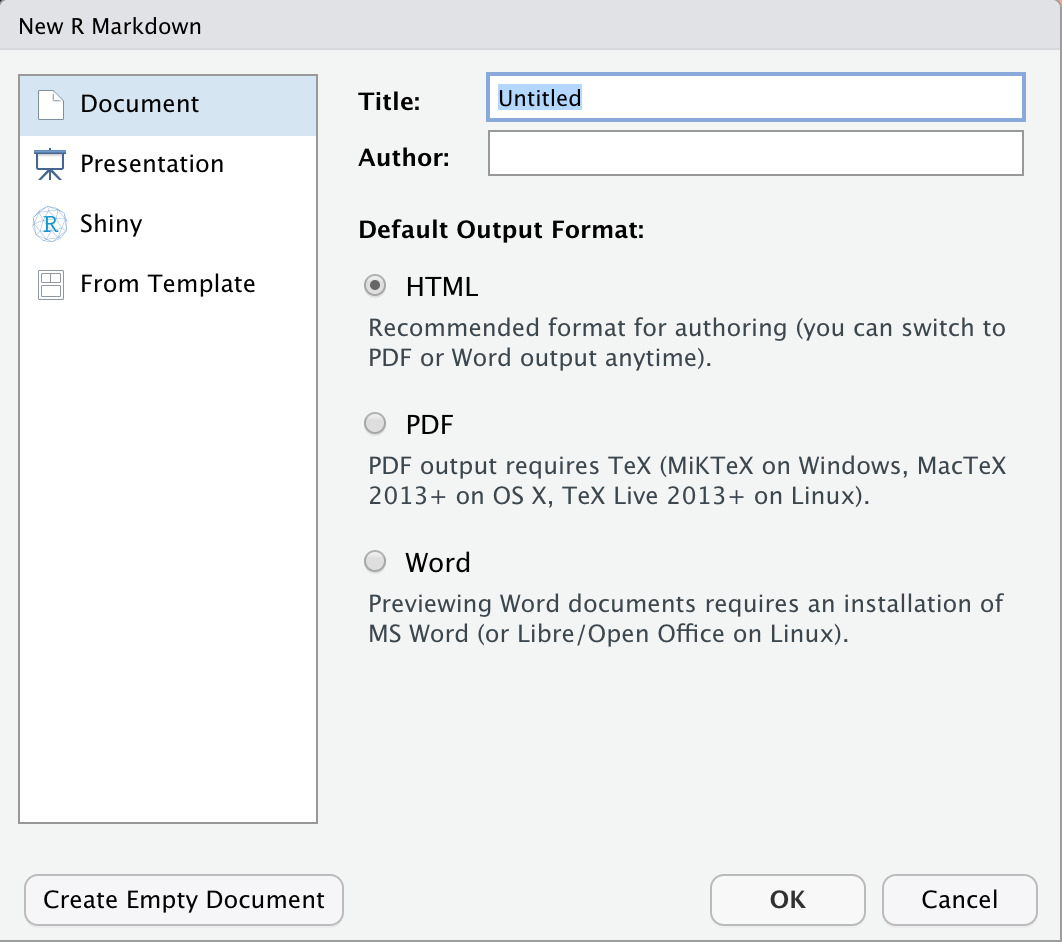
Figure 3: New R Markdown file
Use the default settings for HTML (we can always change it later), and click OK.
Once the Rmarkdown document appears and opens in RStudio, we should see something like this:

Figure 4: The blank R Markdown template
If you look at the top right of the code editor window, there’s a small “A” shaped compass1.

If you click on that icon, the R Markdown document will render into something that is visually more appealing and essentially will look much more like a word processor might.

Figure 5: Visual markdown editor
yaml header
yaml stands for
yet another markup language. Save that for a trivia
question in the future. Every Rmarkdown document uses a
yaml header (the bits between the --- marks)
at the top of the document to tell your computer how R should knit your
document together (stitch all the different parts together into a single
seamless document).
There are many possible options here and they can depend on the type of document we want to create, but typically the key pieces are:
title: Name of our documentoutput: what we end up knitting our R markdown into (e.g.,.pdf,.html,.docx). See the RStudio R Markdown website for more details on different format options.
Let’s focus on making an .html file, and add some
parameters about the table of contents. We can add
toc: true and toc_float: true to our yaml so
it looks like this:
---
title: "Untitled"
output:
html_document:
toc: true
toc_float: true
---Now each section and subsection of our document will appear in the table of contents. How do we make a section or subsection? Read on!
Body Text
The body of our document is typically text written in Markdown2. Markdown is a simple text language that helps make it very easy to just sit down and type without worrying about formatting, and it works across many different operating systems and applications. There are a few basic formatting options to do things like make font bold, italicized, add numbered or bulleted lists, or add section headers. To learn more, while in RStudio, go to the menu and locate Help at the top of the screen. Under Help > Markdown Quick Reference you’ll find a handy “cheatsheet” to help learn these options. There are many additional RStudio cheatsheets for many topics here, and a specific pdf on using R Markdown is available as a pdf here.
For example, to add a figure using Markdown, we can use the following:

code chunks
The third and final component of an R Markdown document is what makes
it R Markdown. The inclusion of
code! What’s great is while the default is
typically R code, there are quite a few additional code
language options that can be stitched together. Look for the little
+C icon in green at the top of your R Markdown, or go to
Code > Insert Chunk. The keyboard
shortcut to do this is Ctrl + Alt + i.
Let’s go ahead and create a new R code chunk and look at
some of the chunk options.
```{r}
# my empty code chunk!
```There are many code chunk options we can use. Put your cursor after
the {r, and hit tab. There are tons! If
you want to learn more, check the R
Markdown reference guide. The key options that we want to know:
echo: TRUE or FALSE. If TRUE, whatever code we put in the chunk will be displayed when we knit the document.eval: TRUE or FALSE. If TRUE, the code inside the chunk will be evaluated or run.include: TRUE or FALSE. If FALSE, the code is still run, but the results won’t be displayed and neither will the code.3fig.cap: A quoted character string, adds captions to figures from code outputs. One caption per chunk.out.width/out.height: A quoted percentage, i.e., “100%”. This controls the size of the image being output from a given code chunk. Very easy and effective for changing graphic output sizes.
Knit Early and Often
One thing that makes writing in R Markdown fun is the ability to get instant feedback. Clicking the Knit button when a section of your document has been updated will (re)-generate the document. We can change the default location the outputs appear by changing the options via the little gear wheel to the right of the knit button. Look for “Preview in Viewer Pane” and select it. From that point on, every time you click knit, most outputs should appear in the Viewer pane of RStudio.
Knitting frequently can be tedious if you have a large document with lots of figures or visuals. However, it’s a great way to learn, and also to ensure things are rendering correctly and successfully.
Customizing Visuals
There’s lots of great visualization materials out there, but there
are a few helpful tips to keep in mind when using images or graphics in
your R Markdown documents. While the default Markdown option does permit
adding images with the  syntax, it doesn’t
permit as much fine control.
If we use a handy function from the {knitr} package, we
can have more more concise control over the size and placement of an
image without having to learn a lot of special code. For example, we can
use either a local file path, or a url! In addition we can add some
arguments that help us specify the size of the image, using the
out.width or out.height parameters. Finally,
we can add a caption with fig.cap. Note, here we want to
hide the code and just show the image, so we use
echo=FALSE.
```{r rivphoto, echo=FALSE, out.width='80%', fig.cap="A tranquil river (photo: R Peek)"}
knitr::include_graphics("images/river_peek.JPG")
```Figure 6: A tranquil river (photo: R Peek)
Similarly, we can include a URL.
```{r delta, echo=FALSE, fig.cap="Sacramento-San Joaquin Delta, color infrared (image: R Pauloo)"}
url <- "https://raw.githubusercontent.com/richpauloo/rp/master/static/img/delta_cir_2.png"
knitr::include_graphics(url)
```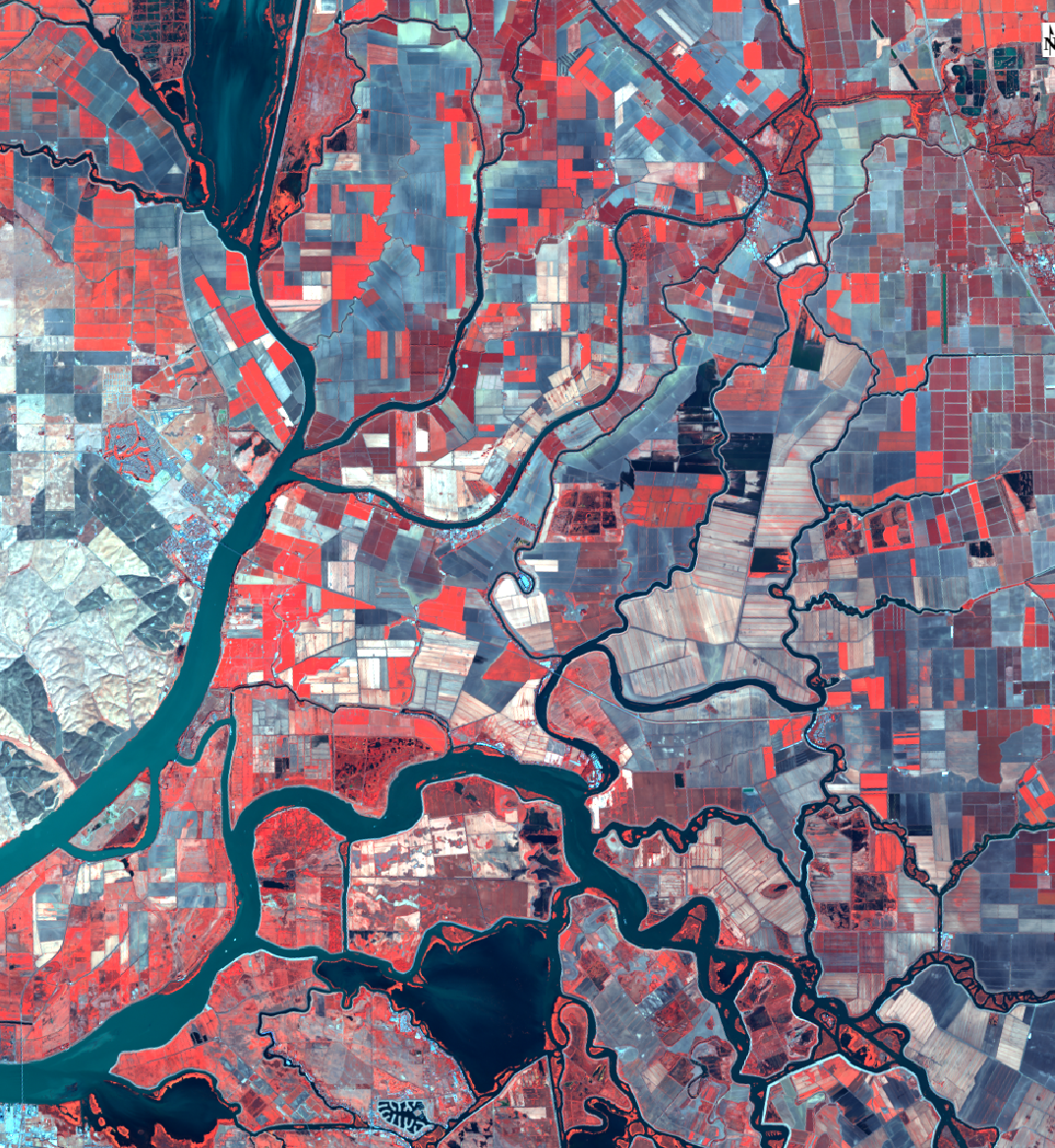
Figure 7: Sacramento-San Joaquin Delta, color infrared (image: R Pauloo)
Tables
Sharing data in tables is a common need for reporting, summary, and analysis. There are a number of packages in R that may be helpful for making tables. This list is not exhaustive, and is meant to just show a few options that may be useful.
- {DT}: Great for interactive and dynamic html based tables
- {kable} & {kableExtra}: Highly customizable static or dynamic tables
- {gt}: Very customizable static or dynamic tables
Let’s load some data we can play with. We’ll use some existing
datasets that come with packages you already have installed. The {dplyr}
package comes with a number of different datasets, as do many R
packages4. We’ll use the
storms dataset since it’s large (+10,000
observations), and our nwis_sites from the American River
which we created in a previous module.
For large unwieldy tables that you may want to just be able to
quickly search or explore, the {DT} package is great,
especially for .html documents. Let’s make a table of the
storms data. A nice feature of the datatable()
is we can search and filter our table interactively.
For a quick and easy static table, the kable() function
from {knitr} is great. We can simply pass a dataframe to
the function and we get a table! For more fancy options, we can use the
functions from the {kableExtra} package.
| name | year | month | day | hour | lat | long | status | category | wind | pressure | tropicalstorm_force_diameter | hurricane_force_diameter |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Amy | 1975 | 6 | 27 | 0 | 27.5 | -79.0 | tropical depression | -1 | 25 | 1013 | NA | NA |
| Amy | 1975 | 6 | 27 | 6 | 28.5 | -79.0 | tropical depression | -1 | 25 | 1013 | NA | NA |
| Amy | 1975 | 6 | 27 | 12 | 29.5 | -79.0 | tropical depression | -1 | 25 | 1013 | NA | NA |
| Amy | 1975 | 6 | 27 | 18 | 30.5 | -79.0 | tropical depression | -1 | 25 | 1013 | NA | NA |
| Amy | 1975 | 6 | 28 | 0 | 31.5 | -78.8 | tropical depression | -1 | 25 | 1012 | NA | NA |
| Amy | 1975 | 6 | 28 | 6 | 32.4 | -78.7 | tropical depression | -1 | 25 | 1012 | NA | NA |
| Amy | 1975 | 6 | 28 | 12 | 33.3 | -78.0 | tropical depression | -1 | 25 | 1011 | NA | NA |
| Amy | 1975 | 6 | 28 | 18 | 34.0 | -77.0 | tropical depression | -1 | 30 | 1006 | NA | NA |
| Amy | 1975 | 6 | 29 | 0 | 34.4 | -75.8 | tropical storm | 0 | 35 | 1004 | NA | NA |
| Amy | 1975 | 6 | 29 | 6 | 34.0 | -74.8 | tropical storm | 0 | 40 | 1002 | NA | NA |
Here’s a slightly fancier option using {kableExtra}. See
the great vignette
for more details.
library(kableExtra)
storms %>%
slice(1:10) %>% # take first 10 rows
kbl() %>%
kable_paper("hover", full_width = T)
| name | year | month | day | hour | lat | long | status | category | wind | pressure | tropicalstorm_force_diameter | hurricane_force_diameter |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Amy | 1975 | 6 | 27 | 0 | 27.5 | -79.0 | tropical depression | -1 | 25 | 1013 | NA | NA |
| Amy | 1975 | 6 | 27 | 6 | 28.5 | -79.0 | tropical depression | -1 | 25 | 1013 | NA | NA |
| Amy | 1975 | 6 | 27 | 12 | 29.5 | -79.0 | tropical depression | -1 | 25 | 1013 | NA | NA |
| Amy | 1975 | 6 | 27 | 18 | 30.5 | -79.0 | tropical depression | -1 | 25 | 1013 | NA | NA |
| Amy | 1975 | 6 | 28 | 0 | 31.5 | -78.8 | tropical depression | -1 | 25 | 1012 | NA | NA |
| Amy | 1975 | 6 | 28 | 6 | 32.4 | -78.7 | tropical depression | -1 | 25 | 1012 | NA | NA |
| Amy | 1975 | 6 | 28 | 12 | 33.3 | -78.0 | tropical depression | -1 | 25 | 1011 | NA | NA |
| Amy | 1975 | 6 | 28 | 18 | 34.0 | -77.0 | tropical depression | -1 | 30 | 1006 | NA | NA |
| Amy | 1975 | 6 | 29 | 0 | 34.4 | -75.8 | tropical storm | 0 | 35 | 1004 | NA | NA |
| Amy | 1975 | 6 | 29 | 6 | 34.0 | -74.8 | tropical storm | 0 | 40 | 1002 | NA | NA |
For nicely formatted tables and lots of control, the
{gt} package is good. It does take a little more wrangling
to get things formatted, but there’s great documentation for this
package as well.
library(gt)
tab1 <- nwis_sites %>%
select(-sourceName) %>%
slice(1:10) %>% # get just first 10 rows
gt()
tab2 <- tab1 %>%
cols_label(identifier = "NWIS ID",
comid = "COMID",
X = "Longitude",
Y = "Latitude") %>%
tab_header(
title = "NWIS Sites on the American River",
subtitle = "Downstream of Nimbus, Sacramento County") %>%
fmt_number(
columns = vars(identifier, comid),
decimals = 0, use_seps = FALSE)
tab2
| NWIS Sites on the American River | |||
| Downstream of Nimbus, Sacramento County | |||
| NWIS ID | COMID | Longitude | Latitude |
|---|---|---|---|
| 11446500 | 948021150 | -121.2277 | 38.63546 |
| 383729121181000 | 15025011 | -121.3038 | 38.62463 |
| 11447540 | 15039173 | -121.5206 | 38.56832 |
| 383438121204200 | 15024993 | -121.3461 | 38.57713 |
| 383609121293200 | 15024919 | -121.4933 | 38.60240 |
| 383052121324401 | 15039157 | -121.5456 | 38.51454 |
| 383205121310901 | 15039157 | -121.5192 | 38.53481 |
| 383457121254900 | 15024941 | -121.4313 | 38.58241 |
| 383515121264400 | 15024941 | -121.4466 | 38.58741 |
| 11446980 | 15024969 | -121.3883 | 38.56713 |
Interactive Maps
And as you’ve seen from previous modules, we can also add maps! This
is also a really nice way to share lots of information. We can only add
{mapview} maps if we are using an html-based output, but
they work well in R Markdown.
Let’s plot our NWIS Stations from the American River that we already
loaded above. First we need to convert these data to {sf}. Let’s show
code that does that. Remember to use eval=TRUE and
echo=TRUE! One additional tip is to label your R code
chunks. This helps us troubleshoot when things are not knitting
properly, and helps keep things organized.
```{r make-nwis-sf, echo=TRUE, eval=TRUE}
library(sf)
# make nwis_sites spatial by converting to sf
nwis_sites_sf <- st_as_sf(nwis_sites, coords=c("X", "Y"), remove=FALSE, crs=4326)
```Next we can make our map with mapview(). Note, there may
be a minor rendering issue with {mapview}. Adding
fgb=FALSE seems to address this!5
Note, here we are going to hide our code (echo = FALSE in
the code chunk options) so that we only show the results!
Extending R Markdown
We’ve demonstrated that R Markdown can be used for creating html
files. This is the tip of the iceberg, and the foundation of building
websites and dashboards with Rmd. Remember
that Rmd files can also knit to
.doc and .pdf files, although we don’t cover
it in this module. Also, because Rmd files
are code, as you level up your R skills, you can begin to automate
generating tens, hundreds, even thousands of reports from a single
template. To extend your understanding of the capabilities of
Rmd, check out One
R Markdown document, 14 demos, a video of a talk by R Markdown
creator and developer Yihui Xie at rstudio::conf(2020).
Additional Resources
There many different types of outputs possible, including slide presentations (see {xaringan} and RStudio’s page on slides), pdfs, and word docs.
Here’s a sample of some good resources that are freely available online:
- One
R Markdown document, 14 demos: a video of a talk by R Markdown
creator and developer Yihui Xie at rstudio::conf(2020)
- R Markdown (online book): all the tips and details you could ever want
- Tips for Working with Images in R Markdown: Helpful tips on sizing and image resolution
- Working with pdf’s via R Markdown using the {tinytex} package: you’ll need a LaTeX distribution installed in order to render or knit to pdf. The {tinytex} package is a great solution.
Previous
module:
12. EDA
Next
module:
14. Troubleshooting
Read more on the simple markup language Markdown here.↩︎
Code that has been evaluated in one chunk can be referred to or used in subsequent chunks. So you can load objects at the start of your document and use them throughout the entire document.↩︎
To figure out what datasets are available across all installed packages, try
data(package = .packages(all.available = TRUE)).↩︎{mapview} is not a static package. Rather, it’s under active development. At the time of writing, the solution to inserting mapview objects into an html document is to use
mapviewOptions(fgb=FALSE). In the future, this may change so that it’s not needed and mapview objects in an html document “just work”. A benefit of using open source projects is that conversations about these fixes and updates can sometimes come from the package developers themselves, as in the case of this mapview issue discussed on Github that provided our fix.```{r make-nwis-mapview, echo=FALSE, eval=TRUE}
library(mapview) mapviewOptions(fgb=FALSE)
mapview(nwis_sites_sf, col.regions=“cyan4”, layer=“NWIS Sites”)
```↩︎